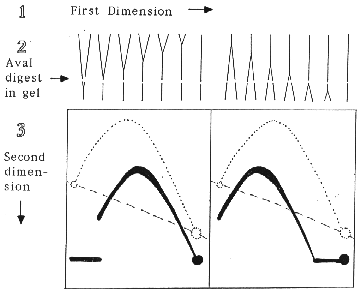
Two dimensional agarose gels have proven to be very useful for identifying restriction fragments that contain replication origins and termini. However, the majority of genomic restriction fragments are replicated by a single fork passing through them. These fragments can be informative if the direction of fork movement can be determined. A simple modification of our 2-D gel procedure, that uses non-denaturing conditions for both dimensions (Brewer and Fangman, 1987, Cell 51:463), permits determination of the direction of fork movement without interference from nicked or broken DNA.
As an example of the procedure, consider an XbaI restriction fragment that is replicated by a single replication fork and has an AvaI site near one end. The XbaI cleaved replication intermediates are separated by mass in the first dimension of electrophoresis. The gel lane is excised, the DNA digested in the gel with AvaI and subjected to the second dimension electrophoresis where separation is by both mass and structure. A Southern blot probed for the larger XbaI-AvaI fragment will show that the detected replication intermediates, while reduced in size, still generate an arc of simple-Ys. However, the position of the 2-D pattern with respect to the arc of full-length XbaI fragments depends upon whether AvaI cleavage removes the first or the last end of the fragment to be replicated. The expected results are illustrated below.
Procedure for restriction enzyme cleavage in gelo. Adapted from the procedure published in the New England Biolabs Catalog (1988-1989; p. 142). The volumes and times recommended here are not necessarily optimal, since I haven't tried reducing them. Note that some restriction enzymes are inhibited by the presence of contaminants in agarose. As a trial run, I recommend cutting lambda DNA, for example, with the enzyme of choice in various brands of agarose. The enzymes that cut to completion in Beckman LE agarose are AvaI, Bam HI, BglII, EcoRV, NcoI, PstI, PvuII, SacII, SnaBI, SpeI, StuI, XbaI and XhoI. (There are likely to be many others that also cut to completion--see The NEB Transcript, Vol. 3 No. 1, March, 1991, pg 8-9.)
1. After staining and photographing the first dimension gel, excise the lane and place it in a disposable pipette reagent reservoir from Costar (#4870; Cambridge MA). These plastic boats have a wedge shaped reservoir that is 12 cm long. Fill up the container with 10 mM Tris (pH 8.0), 0.1 mM EDTA. Incubate at room temperature for 30 minutes with gentle agitation. Repeat this step once.
2. Make up 250 ml of restriction enzyme buffer according to the manufacturer's recipe. Be sure to include BSA but do not include spermidine in the buffer as it might alter the mobility of the DNA in the second dimension. Drain the Tris-EDTA from the gel piece and fill the container with the restriction buffer. Incubate 1 hr at room temperature, with gentle agitation. Repeat this step once.
3. Drain the restriction buffer from the gel slice. Remove excess buffer with a pasteur pipette and blot the gel slab with a kimwipe. Pipette directly onto the surface of the gel, 10 to 20 microliters of enzyme. Seal the reservoir with parafilm and incubate at 37o. (Alternatively, set the reservoir in a glass blaking dish with water in the bottom and cover with saran wrap.) Digest 4 hours (or more).
4. Fill the reservoir with 10 mM Tris, 1 mM EDTA. Agitate gently at room temperature 30 minutes.
5. The gel is now ready for the second dimension of electrophoresis. Beware that the DNA fragments are now smaller and thus the second dimension may need to be a higher concentration of agarose and/or may need to be run for a shorter time. The ethidium bromide stained profile may still contain a prominant arc of linears in the smaller size range; the larger fragments are more likely to have a site for the second enzyme and thus they will be distributed in a haze below the arc of linears. When analyzing the yeast genome it is possible to see single copy sequences as individual spots in this portion of the gel. In addition, the repeated sequences (rDNA and 2 micron) can provide a visual test for the apparent completeness of the second digest.
日前,国产期刊TheInnovation获得首个影响因子(IF=32.1),成为科睿唯安JCR综合性期刊分类下排名仅次于《自然》(IF=64.8)和《科学》(IF=56.9)的期刊,并且这本期刊在目前......
近日,服务科学领域的全球领导者赛默飞世尔科技(以下简称赛默飞)宣布,在达成收购意向两个月之后,赛默飞以28亿美元、折合人民币约190亿元的价格,完成了对TheBindingSiteGroup的全现金收......
11月15日,施普林格·自然和TheLens平台宣布结成重要的合作伙伴关系,以更深入地揭示学术研究和数据如何能通过经济和社会成效,加速推动创新的问题解决方式。通过将科学、投资和企业领域的开放数据更好地......
蛋白质作为构成人体组织器官的支架和主要物质,在人体生命活动中起着重要作用。蛋白质的相互作用能产生许多效应,如形成特异底物作用通道、生成新的结合位点、失活、作用底物专一性和动力学变化等,细胞的代谢、信号......
2021年9月9日,无锡臻和生物科技有限公司(以下简称“臻和科技”)与美国VyantBio公司签署TissueofOrigin®(以下简称“TOO®”)全球权益和ZL转让协议,全资收购这款唯一获FDA......
2021年7月20日,JournalofCellularPhysiology及JournalofCellularBiochemistry同时撤回了中国学者49篇文章。从2019年开始,Journalo......
6月10日,QS教育集团正式发布了2021年世界大学排名,中国共有83所高校上榜,包括内地高校51所,港澳台地区高校32所。中国大学的总体排名情况已经连续数年呈上升趋势,今年再度刷新了榜单。大学排名,......
磷酸甘油酸突变酶1(PGAM1)通过其代谢活性以及与其他蛋白质(例如α平滑肌肌动蛋白(ACTA2))的相互作用,在癌症代谢和肿瘤进展中起关键作用。变构调节被认为是发现针对PGAM1的高选择性和有效抑制......
作为一种重要的植物激素,茉莉酸不仅调控植物对于机械损伤、昆虫取食和腐生型病原菌侵害的防御反应,还参与调控诸多生长发育过程。basicHelix-Loop-Helix(bHLH)类型转录因子MYC2是茉......
ThePlantCell是植物领域的著名学术期刊,对植物学的发展起到了重要的引领作用。为庆祝创刊30周年,ThePlantCell杂志社邀请部分编委会成员及其他科学家对发表在该杂志的重要研究工作进行评......